The application of Bayesian hierarchical models to quantify individual diet specialization
Abstract
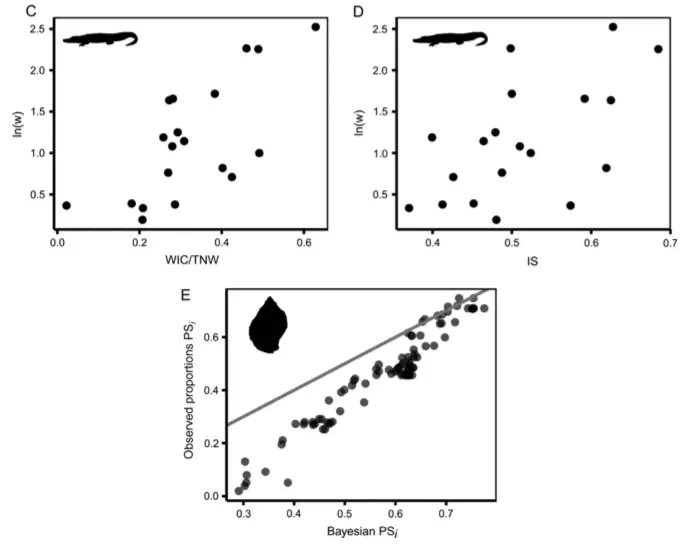
Intraspecific variation in ecologically relevant traits is widespread. In generalist predators in particular, individual diet specialization is likely to have important consequences for food webs. Understanding individual diet specialization empirically requires the ability to quantify individual diet preferences accurately. Here we compare the currently used frequentist maximum likelihood approach, which infers individual preferences using the observed prey proportions to Bayesian hierarchical models that instead estimate these proportions. Using simulated and empirical data, we find that the approach of using observed prey proportions consistently overestimates diet specialization relative to the Bayesian hierarchical approach when the number of prey observations per individual is low or the number of prey observations vary among individuals, two common features of empirical data. Furthermore, the Bayesian hierarchical approach permits the estimation of point estimates for both prey proportions and their variability within and among levels of organization (i.e., individuals, experimental treatments, populations), while also characterizing the uncertainty of these estimates in ways inaccessible to frequentist methods. The Bayesian hierarchical approach provides a useful framework for improving the quantification and understanding of intraspecific variation in diet specialization studies.